Idealized Synthetic Data
Under development
import numpy as np
import pandas as pd
import xarray as xr
from IPython.display import display # so can run as script too
from melodies_monet import driver
Please install h5py to open files from the Amazon S3 servers.
Please install h5netcdf to open files from the Amazon S3 servers.
an = driver.analysis()
an.control = "control_idealized.yaml"
an.read_control()
an
analysis(
control='control_idealized.yaml',
control_dict=...,
models={},
obs={},
paired={},
start_time=Timestamp('2019-09-09 00:00:00'),
end_time=Timestamp('2019-09-10 00:00:00'),
time_intervals=None,
download_maps=True,
output_dir='./output/idealized',
output_dir_save='./output/idealized',
output_dir_read='./output/idealized',
debug=True,
save={'paired': {'method': 'netcdf', 'prefix': 'asdf', 'data': 'all'}},
read={'paired': {'method': 'netcdf', 'filenames': {'test_obs_test_model': 'asdf_test_obs_test_model.nc4'}}},
)
Note: This is the complete file that was loaded.
control_idealized.yaml
1analysis:
2 start_time: "2019-09-09 00:00"
3 end_time: "2019-09-10 00:00"
4 output_dir: ./output/idealized
5 # output_dir_save: # defaults to `output_dir`
6 # output_dir_read: # defaults to `output_dir`
7 debug: True
8 save:
9 paired:
10 method: 'netcdf' # 'netcdf' or 'pkl'
11 prefix: 'asdf' # use only with method=netcdf; don't set if you don't want a fn prefix
12 # output_name: '0905.pkl' # use only with method=pkl
13 data: 'all'
14 # ^ 'all' to save out all pairs or
15 # ['pair1','pair2',...] to save out specific pairs.
16 # With method='pkl' this is ignored and always saves all.
17 # models:
18 # obs:
19 read:
20 paired:
21 method: 'netcdf' # 'netcdf' or 'pkl'
22 filenames:
23 test_obs_test_model: 'asdf_test_obs_test_model.nc4'
24 # filenames: ['0904.pkl','0905.pkl'] # example for pkl method, uses str or iterable of filenames
25 # models:
26 # obs:
27
28model:
29 test_model:
30 files: test_model.nc
31 mod_type: random
32 variables:
33 A:
34 units: "Units of A"
35 unit_scale: 1
36 unit_scale_method: "*"
37 B:
38 units: "Units of B"
39 unit_scale: 1
40 unit_scale_method: "*"
41 mapping:
42 test_obs:
43 A: "A_obs"
44 B: "B_obs"
45 projection: ~ # unused
46
47obs:
48 test_obs:
49 # use_airnow: True
50 filename: test_obs.nc
51 obs_type: pt_sfc
52
53plots:
54 plot_grp1:
55 type: 'timeseries'
56 default_plot_kwargs: # required (with at least one key)
57 linewidth: 2.0
58 domain_type: ['all'] # required
59 domain_name: ['CONUS'] # required
60 data: ['test_obs_test_model'] # required
61 data_proc: # These four seem to be required for time series
62 rem_obs_nan: True # True: Remove all points where model or obs variable is NaN. False: Remove only points where model variable is NaN.
63 ts_select_time: 'time' # 'time' for UTC or 'time_local'
64 ts_avg_window: '3H' # Options: None for no averaging or list pandas resample rule (e.g., 'H', 'D')
65 # ^ TODO: null setting seems not working
66 set_axis: False # If True, add vmin_plot and vmax_plot for each variable in obs.
67
68 plot_grp2:
69 type: 'spatial_overlay'
70 fig_kwargs:
71 states: True
72 figsize: [10, 5]
73 domain_type: ['all'] # required
74 domain_name: ['CONUS'] # required
75 data: ['test_obs_test_model'] # required
76 data_proc:
77 rem_obs_nan: True
78 set_axis: True
Generate data
Model
Show code cell source
rs = np.random.RandomState(42)
control = an.control_dict
nlat = 100
nlon = 200
lon = np.linspace(-161, -60, nlon)
lat = np.linspace(18, 60, nlat)
Lon, Lat = np.meshgrid(lon, lat)
time = pd.date_range(control['analysis']['start_time'], control['analysis']['end_time'], freq="3H")
ntime = time.size
# Generate translating and expanding Gaussian
x_ = np.linspace(-1, 1, lon.size)
y_ = np.linspace(-1, 1, lat.size)
x, y = np.meshgrid(x_, y_)
mu = np.linspace(-0.5, 0.5, ntime)
sigma = np.linspace(0.3, 1, ntime)
g = np.exp(
-(
(
(x[np.newaxis, ...] - mu[:, np.newaxis, np.newaxis])**2
+ y[np.newaxis, ...]**2
) / (
2 * sigma[:, np.newaxis, np.newaxis]**2
)
)
)
# Coordinates
lat_da = xr.DataArray(lat, dims="lat", attrs={'longname': 'latitude', 'units': 'degN'}, name="lat")
lon_da = xr.DataArray(lon, dims="lon", attrs={'longname': 'longitude', 'units': 'degE'}, name="lon")
time_da = xr.DataArray(time, dims="time", name="time")
# Generate dataset
field_names = control['model']['test_model']['variables'].keys()
ds_dict = dict()
for field_name in field_names:
units = control['model']['test_model']['variables'][field_name]['units']
# data = rs.rand(ntime, nlat, nlon)
data = g
da = xr.DataArray(
data,
# coords={"lat": lat_da, "lon": lon_da, "time": time_da},
coords=[time_da, lat_da, lon_da],
dims=['time', 'lat', 'lon'],
attrs={'units': units},
)
ds_dict[field_name] = da
ds = xr.Dataset(ds_dict).expand_dims("z", axis=1)
ds["z"] = [1]
ds_mod = ds
ds_mod
<xarray.Dataset>
Dimensions: (time: 9, lat: 100, lon: 200, z: 1)
Coordinates:
* time (time) datetime64[ns] 2019-09-09 2019-09-09T03:00:00 ... 2019-09-10
* lat (lat) float64 18.0 18.42 18.85 19.27 ... 58.73 59.15 59.58 60.0
* lon (lon) float64 -161.0 -160.5 -160.0 -159.5 ... -61.02 -60.51 -60.0
* z (z) int32 1
Data variables:
A (time, z, lat, lon) float64 0.000964 0.001019 ... 0.5379 0.5353
B (time, z, lat, lon) float64 0.000964 0.001019 ... 0.5379 0.5353ds.squeeze("z").A.plot(col="time", col_wrap=5, size=3);
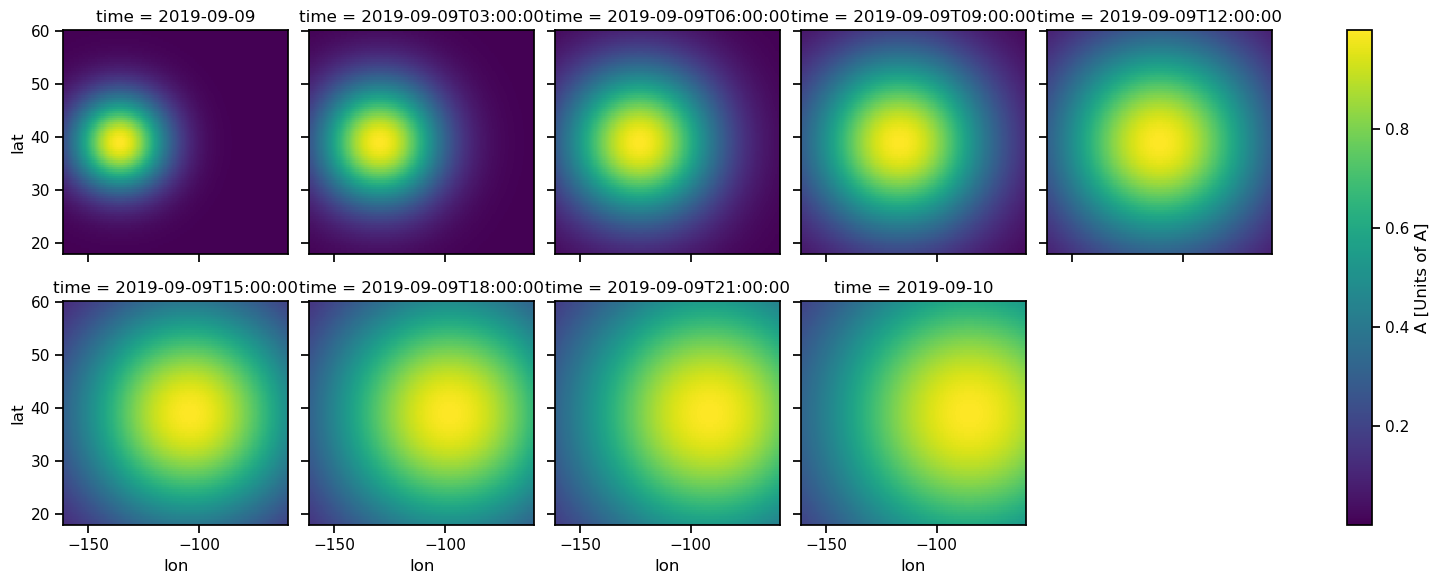
ds.to_netcdf(control['model']['test_model']['files'])
Obs
Show code cell source
from string import ascii_lowercase
# Generate positions
# TODO: only within land boundaries would be cleaner
n = 500
lats = rs.uniform(lat[0], lat[-1], n)
lons = rs.uniform(lon[0], lon[-1], n)
siteid = np.array([f"{x:0{len(str(n))}}" for x in range(n)])[np.newaxis, :]
something_vlen = np.array(
[
"".join(rs.choice(list(ascii_lowercase), size=x, replace=True))
for x in rs.randint(low=2, high=8, size=n)
],
dtype=str,
)[np.newaxis, :]
# Generate dataset
field_names = control['model']['test_model']['variables'].keys()
ds_dict = dict()
for field_name0 in field_names:
field_name = control['model']['test_model']['mapping']['test_obs'][field_name0]
units = control['model']['test_model']['variables'][field_name0]['units']
values = (
ds_mod.A.squeeze().interp(lat=xr.DataArray(lats), lon=xr.DataArray(lons)).values
+ rs.normal(scale=0.3, size=(ntime, n))
)[:, np.newaxis]
da = xr.DataArray(
values,
coords={
"x": ("x", np.arange(n)), # !!!
"time": ("time", time),
"latitude": (("y", "x"), lats[np.newaxis, :], lat_da.attrs),
"longitude": (("y", "x"), lons[np.newaxis, :], lon_da.attrs),
"siteid": (("y", "x"), siteid),
"something_vlen": (("y", "x"), something_vlen),
},
dims=("time", "y", "x"),
attrs={'units': units},
)
ds_dict[field_name] = da
ds = xr.Dataset(ds_dict)
ds
<xarray.Dataset>
Dimensions: (x: 500, time: 9, y: 1)
Coordinates:
* x (x) int32 0 1 2 3 4 5 6 7 ... 493 494 495 496 497 498 499
* time (time) datetime64[ns] 2019-09-09 ... 2019-09-10
latitude (y, x) float64 33.73 57.93 48.74 43.14 ... 21.26 58.92 59.42
longitude (y, x) float64 -90.49 -106.9 -129.7 ... -147.2 -65.03 -116.0
siteid (y, x) <U3 '000' '001' '002' '003' ... '497' '498' '499'
something_vlen (y, x) <U7 'xicjs' 'cvoaglq' 'uzxt' ... 'dzzfatw' 'poqhzl'
Dimensions without coordinates: y
Data variables:
A_obs (time, y, x) float64 0.21 0.1793 0.1735 ... 0.118 0.6552
B_obs (time, y, x) float64 -0.1099 -0.2269 ... 0.1155 0.6365ds.to_netcdf(control['obs']['test_obs']['filename'])
Load
an.open_models()
test_model.nc
**** Reading Unspecified model output. Take Caution...
an.models['test_model'].obj
Show code cell output
<xarray.Dataset>
Dimensions: (time: 9, lat: 100, lon: 200, z: 1)
Coordinates:
* time (time) datetime64[ns] 2019-09-09 2019-09-09T03:00:00 ... 2019-09-10
* lat (lat) float64 18.0 18.42 18.85 19.27 ... 58.73 59.15 59.58 60.0
* lon (lon) float64 -161.0 -160.5 -160.0 -159.5 ... -61.02 -60.51 -60.0
* z (z) int32 1
Data variables:
A (time, z, lat, lon) float64 0.000964 0.001019 ... 0.5379 0.5353
B (time, z, lat, lon) float64 0.000964 0.001019 ... 0.5379 0.5353an.open_obs()
an.obs['test_obs'].obj
Show code cell output
<xarray.Dataset>
Dimensions: (x: 500, time: 9, y: 1)
Coordinates:
* x (x) int32 0 1 2 3 4 5 6 7 ... 493 494 495 496 497 498 499
* time (time) datetime64[ns] 2019-09-09 ... 2019-09-10
latitude (y, x) float64 ...
longitude (y, x) float64 ...
siteid (y, x) object ...
something_vlen (y, x) object ...
Dimensions without coordinates: y
Data variables:
A_obs (time, y, x) float64 ...
B_obs (time, y, x) float64 ...Pair
%%time
an.pair_data()
Show code cell output
After pairing: time latitude longitude siteid something_vlen \
0 2019-09-09 00:00:00 33.730685 -90.485667 000 xicjs
1 2019-09-09 03:00:00 33.730685 -90.485667 000 xicjs
2 2019-09-09 06:00:00 33.730685 -90.485667 000 xicjs
3 2019-09-09 09:00:00 33.730685 -90.485667 000 xicjs
4 2019-09-09 12:00:00 33.730685 -90.485667 000 xicjs
... ... ... ... ... ...
4495 2019-09-09 12:00:00 59.420851 -115.953417 499 poqhzl
4496 2019-09-09 15:00:00 59.420851 -115.953417 499 poqhzl
4497 2019-09-09 18:00:00 59.420851 -115.953417 499 poqhzl
4498 2019-09-09 21:00:00 59.420851 -115.953417 499 poqhzl
4499 2019-09-10 00:00:00 59.420851 -115.953417 499 poqhzl
A_obs B_obs A B
0 0.209983 -0.109890 0.008033 0.008033
1 0.171024 0.289535 0.111157 0.111157
2 -0.174732 0.634226 0.343373 0.343373
3 0.518397 0.673546 0.587818 0.587818
4 1.091830 1.225345 0.769533 0.769533
... ... ... ... ...
4495 0.160357 0.926215 0.316865 0.316865
4496 0.652020 0.369675 0.394017 0.394017
4497 0.713842 0.143496 0.450187 0.450187
4498 0.328453 0.524407 0.489130 0.489130
4499 0.655239 0.636502 0.515131 0.515131
[4500 rows x 9 columns]
CPU times: total: 109 ms
Wall time: 85 ms
an.paired
{'test_obs_test_model': pair(
type='pt_sfc',
radius_of_influence=1000000.0,
obs='test_obs',
model='test_model',
model_vars=['A', 'B'],
obs_vars=['A_obs', 'B_obs'],
filename='test_obs_test_model.nc',
)}
an.paired['test_obs_test_model'].obj
<xarray.Dataset>
Dimensions: (time: 9, x: 500)
Coordinates:
* time (time) datetime64[ns] 2019-09-09 ... 2019-09-10
* x (x) int32 0 1 2 3 4 5 6 7 ... 493 494 495 496 497 498 499
Data variables:
something_vlen (time, x) object 'xicjs' 'cvoaglq' ... 'dzzfatw' 'poqhzl'
A_obs (time, x) float64 0.21 0.1793 0.1735 ... 0.462 0.118 0.6552
B_obs (time, x) float64 -0.1099 -0.2269 0.05157 ... 0.1155 0.6365
A (time, x) float64 0.008033 0.001784 0.2917 ... 0.5939 0.5151
B (time, x) float64 0.008033 0.001784 0.2917 ... 0.5939 0.5151
latitude (x) float64 33.73 57.93 48.74 43.14 ... 21.26 58.92 59.42
longitude (x) float64 -90.49 -106.9 -129.7 ... -147.2 -65.03 -116.0
siteid (x) object '000' '001' '002' '003' ... '497' '498' '499'an.paired['test_obs_test_model'].obj.dims
Frozen({'time': 9, 'x': 500})
Plot
%%time
an.plotting()
Warning: variables dict for A_obs not provided, so defaults used
C:\Users\zmoon\git\MELODIES-MONET\melodies_monet\plots\surfplots.py:671: FutureWarning: The default value of numeric_only in DataFrameGroupBy.mean is deprecated. In a future version, numeric_only will default to False. Either specify numeric_only or select only columns which should be valid for the function.
df_mean=df.groupby(['siteid'],as_index=False).mean()
Warning: variables dict for B_obs not provided, so defaults used
C:\Users\zmoon\git\MELODIES-MONET\melodies_monet\plots\surfplots.py:671: FutureWarning: The default value of numeric_only in DataFrameGroupBy.mean is deprecated. In a future version, numeric_only will default to False. Either specify numeric_only or select only columns which should be valid for the function.
df_mean=df.groupby(['siteid'],as_index=False).mean()
CPU times: total: 5.3 s
Wall time: 5.31 s
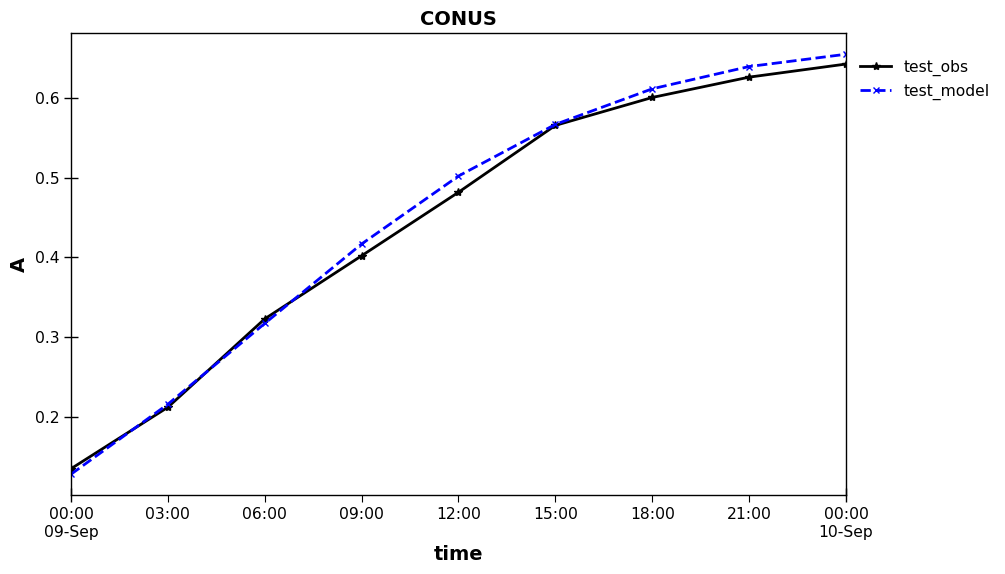
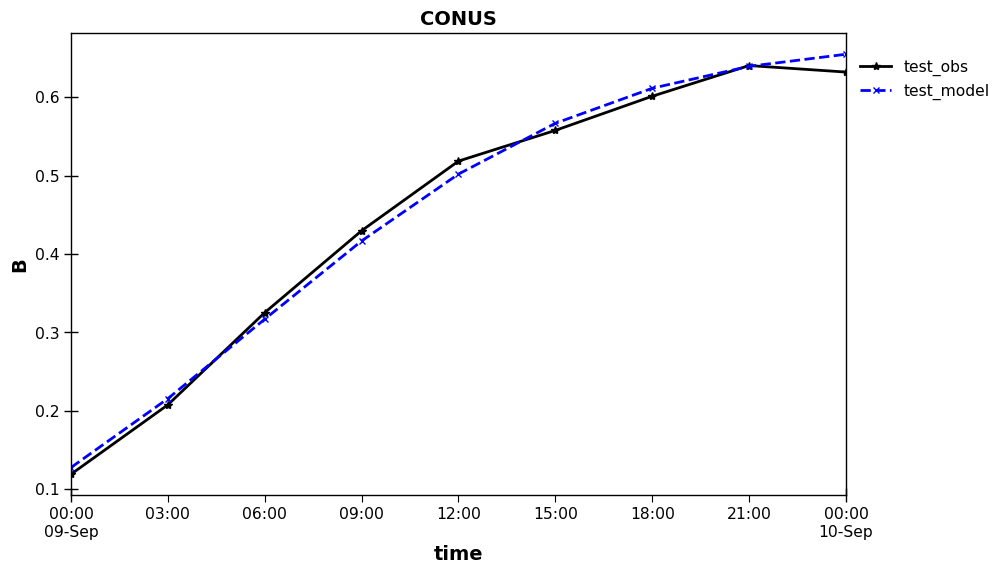
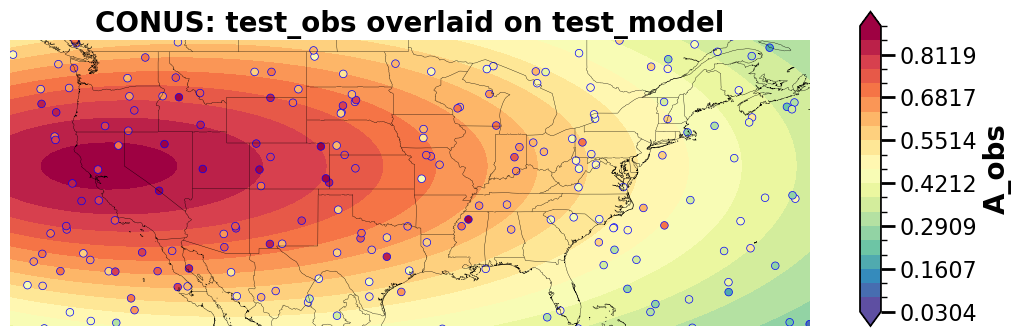
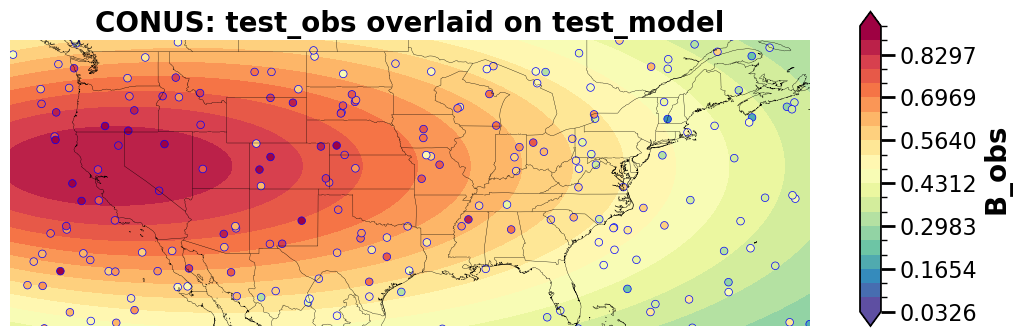
Save/load paired data – netCDF
And compare to the original pair object.
from copy import deepcopy
p0 = deepcopy(an.paired['test_obs_test_model'].obj)
an.save_analysis()
an.read_analysis()
p1 = deepcopy(an.paired['test_obs_test_model'].obj)
p1.close()
display(p0)
display(p1)
assert p1 is not p0 and p1.equals(p0)
Writing: ./output/idealized\asdf_test_obs_test_model.nc4
Reading: ./output/idealized\asdf_test_obs_test_model.nc4
<xarray.Dataset>
Dimensions: (time: 9, x: 500)
Coordinates:
* time (time) datetime64[ns] 2019-09-09 ... 2019-09-10
* x (x) int32 0 1 2 3 4 5 6 7 ... 493 494 495 496 497 498 499
Data variables:
something_vlen (time, x) object 'xicjs' 'cvoaglq' ... 'dzzfatw' 'poqhzl'
A_obs (time, x) float64 0.21 0.1793 0.1735 ... 0.462 0.118 0.6552
B_obs (time, x) float64 -0.1099 -0.2269 0.05157 ... 0.1155 0.6365
A (time, x) float64 0.008033 0.001784 0.2917 ... 0.5939 0.5151
B (time, x) float64 0.008033 0.001784 0.2917 ... 0.5939 0.5151
latitude (x) float64 33.73 57.93 48.74 43.14 ... 21.26 58.92 59.42
longitude (x) float64 -90.49 -106.9 -129.7 ... -147.2 -65.03 -116.0
siteid (x) object '000' '001' '002' '003' ... '497' '498' '499'<xarray.Dataset>
Dimensions: (time: 9, x: 500)
Coordinates:
* time (time) datetime64[ns] 2019-09-09 ... 2019-09-10
* x (x) int32 0 1 2 3 4 5 6 7 ... 493 494 495 496 497 498 499
Data variables:
something_vlen (time, x) object ...
A_obs (time, x) float64 ...
B_obs (time, x) float64 ...
A (time, x) float64 ...
B (time, x) float64 ...
latitude (x) float64 ...
longitude (x) float64 ...
siteid (x) object ...
Attributes:
title:
format: NetCDF-4
date_created: 2023-02-27
dict_json: {\n "type": "pt_sfc",\n "radius_of_influence": 10000...
group_name: test_obs_test_modelSave/load paired data – Python object
print(an.save)
an.save["paired"]["method"] = "pkl"
del an.save["paired"]["prefix"]
# We could leave `prefix` since unused, but we need to set `output_name`
an.save["paired"]["output_name"] = "asdf.joblib"
print("->", an.save)
print()
print(an.read)
an.read["paired"]["method"] = "pkl"
an.read["paired"]["filenames"] = "asdf.joblib"
print("->", an.read)
print()
an.save_analysis()
an.read_analysis()
p2 = deepcopy(an.paired['test_obs_test_model'].obj)
p2.close()
# display(p0)
display(p2)
assert p2 is not p0 and p2.equals(p0)
{'paired': {'method': 'netcdf', 'prefix': 'asdf', 'data': 'all'}}
-> {'paired': {'method': 'pkl', 'data': 'all', 'output_name': 'asdf.joblib'}}
{'paired': {'method': 'netcdf', 'filenames': {'test_obs_test_model': ['./output/idealized\\asdf_test_obs_test_model.nc4']}}}
-> {'paired': {'method': 'pkl', 'filenames': 'asdf.joblib'}}
Writing: ./output/idealized\asdf.joblib
Reading: ./output/idealized\asdf.joblib
<xarray.Dataset>
Dimensions: (time: 9, x: 500)
Coordinates:
* time (time) datetime64[ns] 2019-09-09 ... 2019-09-10
* x (x) int32 0 1 2 3 4 5 6 7 ... 493 494 495 496 497 498 499
Data variables:
something_vlen (time, x) object ...
A_obs (time, x) float64 ...
B_obs (time, x) float64 ...
A (time, x) float64 ...
B (time, x) float64 ...
latitude (x) float64 ...
longitude (x) float64 ...
siteid (x) object ...
Attributes:
title:
format: NetCDF-4
date_created: 2023-02-27
dict_json: {\n "type": "pt_sfc",\n "radius_of_influence": 10000...
group_name: test_obs_test_model